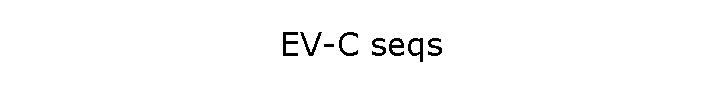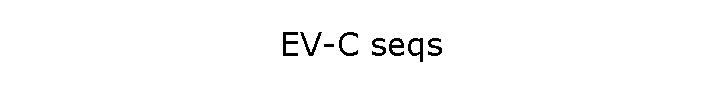|
|
| |
Genus: Enterovirus
Species: Enterovirus C
| Serotype: Coxsackievirus A1 (CVA1) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A11 (CVA11) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A13 (CVA13) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A17 (CVA17) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A19 (CVA19) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A20 (CVA20) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A21 (CVA21) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A22 (CVA22) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Coxsackievirus A24 (CVA24) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C95 (EV-C95) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C96 (EV-C96) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C99 (EV-C99) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C102 (EV-C102) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C104 (EV-C104) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C105 (EV-C105) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C109 (EV-C109) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C113 (EV-C113) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C116 (EV-C116) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C117 (EV-C117) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
|
Serotype: Enterovirus C118 (EV-C118) |
|
| 5' UTR |
VP4 |
VP2 |
VP3 |
VP1 |
2A |
2B |
2C |
3A |
3B |
3C |
3D |
3' UTR |
|
| |
|
|
(White areas indicate genes which haven't yet been
sequenced completely;
grey areas are where only partial sequences exist)
References
| Virus |
Strain |
Region |
Length |
Accession No. |
Reference |
Comments |
Coxsackievirus A1 (CVA1)
|
| CVA1 |
T.T. (Tompkins)* (Coxsackie, NY/47) |
5'NCR(p),1AB(p),3'NCR(p) |
199,210,57 |
X87584 |
Pulli et al., 1995 |
|
|
VP1 |
888 |
AF081293 |
Oberste et al., 1999 |
|
|
CG |
7397 |
AF499635 |
Brown et al., 2003 |
|
| CVA1 |
BBD34 (Bangladesh,
2009) |
CG(p) |
7258 |
KC785529 |
Tokarz et al.,
2013 |
end missing |
| |
|
|
|
|
|
|
Coxsackievirus A11 (CVA11)
|
| CVA11 |
Belgium 1* (Belgium/51) |
5'NCR(p),1AB(p),3D(p),3'NCR(p) |
199,210,387,55 |
X87592 |
Pulli et al., 1995 |
|
|
VP1 |
912 |
AF081301 |
Oberste et al., 1999 |
|
|
CG |
7453 |
AF499636 |
Brown et al., 2003 |
|
| CVA11 |
G-9* (South Africa/50)
(formerly CVA15) |
5'NCR(p),3D(p),3'NCR(p) |
199,387,55 |
X87596 |
Pulli et al., 1995 |
|
| VP2(p) |
? |
? |
Oberste et al., 1998 |
|
|
VP1 |
912 |
AF081305 |
Oberste et al., 1999 |
|
|
CG |
7441 |
AF499638 |
Brown et al., 2003 |
|
|
CG |
7441 |
AF465512 |
Newcombe et
al., 2003 |
|
| |
|
|
|
|
|
|
Coxsackievirus A13 (CVA13)
|
| CVA13 |
Flores* (Mexico/52) |
5'NCR(p),1AB(p),3D(p),3'NCR(p) |
199,210,387,55 |
X87594 |
Pulli et al., 1995 |
|
|
VP1 |
927 |
AF081303 |
Oberste et al., 1999 |
|
|
CG |
7458 |
AF499637 |
Brown et al., 2003 |
|
|
CG |
7458 |
AF465511 |
Newcombe et
al., 2003 |
|
| CVA13 |
G-13* (South Africa/50)
(formerly CVA18) |
5'NCR(p),1AB(p),3D(p),3'NCR(p) |
199,210,387,55 |
X87598 |
Pulli et al., 1995 |
|
| VP2(p) |
? |
? |
Oberste et al., 1998 |
|
|
VP1 |
927 |
AF081307 |
Oberste et al., 1999 |
|
|
CG |
7458 |
AF499640 |
Brown et al., 2003 |
|
|
CG |
7457 |
AF465513 |
Newcombe et
al., 2003 |
|
| CVA13 |
BAN00-10494 |
CG |
7457 |
DQ995639 |
Brown et al., 2009 |
|
| |
|
|
|
|
|
|
Coxsackievirus A17 (CVA17)
|
| CVA17 |
G-12* (South Africa/51) |
5'NCR(p),1AB(p),3D(p),3'NCR(p) |
199,210,387,55 |
X87597 |
Pulli et al., 1995 |
|
|
VP1 |
918 |
AF081306 |
Oberste et al., 1999 |
|
|
CG |
7457 |
AF499639 |
Brown et al., 2003 |
|
| |
|
|
|
|
|
|
Coxsackievirus A19 (CVA19)
|
| CVA19 |
NIH-8663 (Dohi)* (Japan/52) |
3D(p),3'NCR(p) |
387,57 |
X87599 |
Pulli et al., 1995 |
|
| VP2(p) |
? |
? |
Oberste et al., 1998 |
|
|
VP1 |
888 |
AF081308 |
Oberste et al., 1999 |
|
|
CG |
7410 |
AF499641 |
Brown et al., 2003 |
|
| CVA19 |
BBD26 (Bangladesh,
2009) |
CG(p) |
7285 |
KC785531 |
Tokarz et al.,
2013 |
ends missing |
| CVA19 |
BBD66 (Bangladesh,
2009) |
CG(p) |
7364 |
KC785532 |
Tokarz et al.,
2013 |
ends missing |
| |
|
|
|
|
|
|
Coxsackievirus A20 (CVA20)
|
| CVA20 |
IH-35* (New York/55) |
5'NCR(p),1AB(p),3D(p) |
199,210,387 |
X87600 |
Pulli et al., 1995 |
|
|
VP1 |
909 |
AF081309 |
Oberste et al., 1999 |
|
|
CG |
7436 |
AF499642 |
Brown et al., 2003 |
|
| CG |
7436 |
AF465514 |
Newcombe et al., 2003 |
|
| CVA20a |
Tulane 1623 (Louisiana/51) |
5'NCR(p),3D(p) |
200,387 |
X87601 |
Pulli et al., 1995 |
|
| CVA20b |
Cecil P2647 (South Africa/51) |
5'NCR(p),1AB(p),3D(p) |
198,210,387 |
X87602 |
Pulli et al., 1995 |
|
| CVA20 |
BAN01-10618 |
CG |
7441 |
EF015014 |
Brown et al., 2009 |
|
| |
|
|
|
|
|
|
Coxsackievirus A21 (CVA21)
|
| CVA21 |
Coe (California/54) |
CG |
7401 |
D00538 |
Hughes et al., 1989 |
|
| 5' NCR(p) |
260 |
AF068880
AF076999 |
Muir et al., unpublished |
|
| CVA21 |
Kuykendall* (California/52) |
CG |
7406 |
AF546702 |
Brown et al., 2003 |
|
|
CG |
7405 |
AF465515 |
Newcombe et
al., 2003 |
|
| CVA21 |
272101 |
P1 |
2637 |
AY319942 |
Newcombe et
al., 2004 |
|
| CVA21 |
275238 |
P1 |
2637 |
AY319943 |
Newcombe et
al., 2004 |
|
| CVA21 |
272598 |
P1 |
2637 |
AY319944 |
Newcombe et
al., 2004 |
|
| CVA21 |
BAN00-10467 |
CG |
7440 |
EF015031 |
Brown et al., 2009 |
|
| |
|
|
|
|
|
|
Coxsackievirus A22 (CVA22)
|
| CVA22 |
Chulman* (New York/55) |
5'NCR(p),3D(p),3'NCR(p) |
197,387,57 |
X87603 |
Pulli et al., 1995 |
|
| VP2(p) |
? |
? |
Oberste et al., 1998 |
|
|
VP1 |
888 |
AF081310 |
Oberste et al., 1999 |
|
|
CG |
7406 |
AF499643 |
Brown et al., 2003 |
|
| CVA22 |
USA75-10624 |
CG |
7402 |
DQ995648 |
Brown et al., 2009 |
|
| CVA22 |
BBD01 (Bangladesh,
2009) |
CG(p) |
7284 |
KC785528 |
Tokarz et al.,
2013 |
ends missing |
| CVA22 |
BBD58 (Bangladesh,
2009) |
CG(p) |
7265 |
KC785530 |
Tokarz et al.,
2013 |
ends missing |
| |
|
|
|
|
|
|
Coxsackievirus A24 (CVA24)
|
| CVA24 |
Joseph* |
5' NCR(p) |
255 |
AF068881 |
Muir et al., unpublished |
|
| VP2(p) |
? |
? |
Oberste et al., 1998 |
|
|
VP1 |
915 |
AF081311 |
Oberste et al., 1999 |
|
| CVA24' |
DN-19 |
VP2(p) |
? |
? |
Oberste et al., 1998 |
|
|
VP1 |
915 |
AF081347 |
Oberste et al., 1999 |
|
| CVA24v |
EH 24/70 (Singapore/70) |
CG |
7461 |
D90457 |
Supanaranond et al., 1992 |
|
| |
|
|
|
|
|
|
Enterovirus C95 (EV-C95)
|
| EV-C95 |
5-05* |
VP1 |
|
N/A |
Norder et al., unpublished |
|
| EV-C95 |
T08-083/Chad/2008 |
VP1 |
894 |
JX417822 |
Sadeuh-Mba et al.,
2013 |
|
| EV-C95 |
T08-234/Chad/2008 |
VP1 |
894 |
JX417823 |
Sadeuh-Mba et al.,
2013 |
|
| |
|
|
|
|
|
|
Enterovirus C96 (EV-C96)
|
|
EV-C96 |
SVK03-24 |
VP1 |
927 |
EF364404 |
Smura
et al., 2007 |
|
|
3D(p) |
459 |
EF364411 |
Smura
et al., 2007 |
|
| EV-C96 |
10488 |
VP1 |
927 |
AY919472 |
Oberste et al., 2006 |
|
| EV-C96 |
BAN00-10488* |
CG |
7478 |
EF015886 |
Brown et al.,
2009 |
|
| EV-C96 |
10548c |
VP1(p) |
477 |
AY919538 |
Oberste et al., 2006 |
|
| EV-C96 |
FIN04-7 |
CG |
7475 |
FJ751914 |
Smura
et al., 2009 |
|
| EV-C96 |
FIN05-2 |
CG |
7471 |
FJ751915 |
Smura
et al., 2009 |
|
| EV-C96 |
AFP809/GD/CHN/2011 |
CG |
7469 |
KF495604 |
Lu et al.,
2014 |
|
| |
|
|
|
|
|
|
Enterovirus C99 (EV-C99)
|
| EV-C99 |
10461-ban00* |
VP1 |
909 |
AY919447 |
Oberste et al., 2006 |
|
| EV-C99 |
USA-GA84-10636 |
CG |
7447 |
EF555644 |
Brown et al.,
2009 |
|
| EV-C99 |
HT-XEBGH09F/XJ/CHN/2011 |
CG |
7455 |
KF129411 |
Sun et al.,
2013 |
|
| EV-C99 |
KSSC-ALXHH01F/XJ/CHN/2011 |
CG |
7455 |
KF129412 |
Sun et al.,
2013 |
|
| |
|
|
|
|
|
|
Enterovirus C102 (EV-C102)
|
| EV-C102 |
10424-BAN99* |
VP1 |
909 |
AY919409 |
Oberste et al., 2006 |
|
| EV-C102 |
BAN99-10424* |
CG |
7438 |
EF555645 |
Brown et al.,
2009 |
|
| |
|
|
|
|
|
|
Enterovirus C104 (EV-C104)
|
| EV-C104 |
CL-1231094 FL* |
CG(p) |
7229 |
EU840733 |
Tapparel et al.,
2009 |
3' end missing |
| EV-C104 |
AK11 (Japan:Osaka
City, Feb-2011) |
CG |
7408 |
AB686524 |
Kaida et al.,
2012 |
|
| EV-C104 |
Pavia259-7712
(Italy, 2009) |
CG |
7409 |
JX982253 |
Piralla et al.,
2013 |
|
| EV-C104 |
Pavia260-9210
(Italy, 2009) |
CG |
7409 |
JX982254 |
Piralla et al.,
2013 |
|
| EV-C104 |
Pavia263-11230
(Italy, 2009) |
CG |
7409 |
JX982255 |
Piralla et al.,
2013 |
|
| EV-C104 |
Pavia264-16291
(Italy, 2009) |
CG |
7409 |
JX982256 |
Piralla et al.,
2013 |
|
| EV-C104 |
Pavia261-9570
(Italy, 2009) |
CG |
7409 |
JX982257 |
Piralla et al.,
2013 |
|
| EV-C104 |
Pavia68-10804B
(Italy, 2009) |
CG |
7409 |
JX982258 |
Piralla et al.,
2013 |
|
| EV-C104 |
Pavia262-11228 (Italy, 2009) |
CG |
7409 |
JX982259 |
Piralla et al.,
2013 |
|
| EV-C104 |
GAM693 (Gambia,
2009) |
CG(p) |
7067 |
KC785523 |
Tokarz et al.,
2013 |
ends missing |
| EV-C104 |
GAM714 (Gambia,
2009) |
CG(p) |
7332 |
KC785524 |
Tokarz et al.,
2013 |
ends missing |
| |
|
|
|
|
|
|
Enterovirus C105 (EV-C105)
|
| EV-C105 |
TW/NTU07-1841* |
VP1 |
|
N/A |
Cheng et al.,
unpublished |
|
| EV-C105 |
34S/DRC/2010 |
CG |
7299 |
JX514943 |
Lukashev et al., 2012 |
|
| EV-C105 |
PER153 (Peru, 2010) |
CG(p) |
7309 |
JX393302 |
Tokarz et al.,
2012 |
|
| EV-C105 |
BU5 (Burundi, 2010) |
VP1(p) |
323 |
JQ317291 |
Esposito et al.,
2010 |
|
| EV-C105 |
BU77 (Burundi, 2010) |
VP1(p) |
325 |
JQ317292 |
Esposito et al.,
2010 |
|
| EV-C105 |
ROM31 (Romania,
2010) |
VP1(p) |
325 |
JQ317293 |
Esposito et al.,
2010 |
|
| EV-C105 |
CY135/Cyprus/2012 |
5'UTR(p)+VP4+VP2(p) |
1045 |
KF322115 |
Richter et al.,
2013 |
|
| VP1 |
973 |
KF322116 |
Richter et al.,
2013 |
|
| |
|
|
|
|
|
|
Enterovirus C109 (EV-C109)
|
| EV-C109 |
NICA08-4327* |
CG |
7354 |
GQ865517 |
Yozwiak et al.,
2010 |
|
| EV-C109 |
L87/HUN/2007 |
CG |
6912 |
JN900470 |
Pankovics et al.,
2012 |
|
| |
|
|
|
|
|
|
Enterovirus C113 (EV-C113
|
| EV-C113 |
BAN-11609* |
VP1 |
|
n/a |
Oberste et al., unpub. |
|
| EV-C113 |
BAN/2008/14800 |
VP1(p) |
311 |
JX538208 |
Oberste et al., 2013 |
|
| EV-C113 |
BBD-48/Bangladesh/2009 |
CG(p) |
7318 |
KC344833 |
Tokarz et al.,
2013 |
|
| EV-C113 |
BBD-83/Bangladesh/2009 |
CG(p) |
7335 |
KC344834 |
Tokarz et al.,
2013 |
|
| |
|
|
|
|
|
|
Enterovirus C116 (EV-C116)
|
| EV-C116 |
126/Russia/2010* |
CG |
7395 |
JX514942 |
Lukashev et al., 2012 |
|
| |
|
|
|
|
|
|
Enterovirus C117 (EV-C117)
|
| EV-C117 |
LIT22/Vilnius,
Lithuania/2011* |
P1 |
2634 |
JQ446368 |
Daleno et al.,
2012 |
|
| CG |
7363 |
JX262382 |
Daleno et al.,
2012 |
|
| EV-C117 |
BCH096A/China, 2007 |
CG(p) |
7196 |
JX560527 |
Xiang & Wang, unpub. |
|
| |
|
|
|
|
|
|
Enterovirus C118 (EV-C118)
|
| EV-C118 |
ISR10*
(Israel/2011) |
VP1 |
891 |
JQ768163 |
Daleno et al.,
2013a |
|
| EV-C118 |
ISR10* (Israel/2011) |
CG |
7357 |
JX961708 |
Daleno et al.,
2013b |
|
| EV-C118 |
ISR38 (Israel/2011) |
CG |
7357 |
JX961709 |
Daleno et al.,
2013b |
|
| EV-C118 |
PER161 (Peru, 2010) |
CG(p) |
7299 |
JX393301 |
Tokarz et al.,
2012 |
|
| EV-C118 |
CQ5185
(China: Chongqing, 2011) |
CG |
7374 |
JX678288 |
Lu et
al., unpub. |
|
| |
|
|
|
|
|
|
CG, complete genome
(p), partial gene sequence
*, prototype strain |
References
|
|