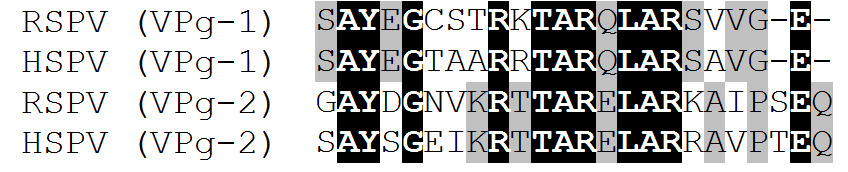The
Picornavirus
Pages
|
Child level |
|
|
|
Same level |
|
|
|
Genera |
|
|
|
Aquamavirus
A is the type species of the genus
Aquamavirus. Seal aquamavirus A1
(formerly seal picornavirus 1) is currently the only
member of the species.
History In 1988, during the seal plaque epidemic in the North Sea (caused by phocine distemper virus), a picorna-like virus was isolated from harbour (common) seals (Phoca vitulina)(Osterhaus, 1988). The virus was found in the lungs of 20 of 22 seals investigated (Osterhaus, 1988). The 3' end of the genome was amplified using a pan-picornavirus RT-PCR and its nucleotide sequence determined (454 nt) (Knowles, 2005). Comparison with all picornavirus sequence known at the time suggested that it belonged to a novel genus. 01/10/2007: The complete genome of "seal picornavirus type 1" (isolate HO.02.21), isolated from Arctic ringed seals (Pusa hispida) in Canada, was released on GenBank (EU142040; Kapoor et al., 2008). Comparison of the sequences obtained by Knowles (2005) and Kapoor et al. (2008) shows a nucleotide identity of 87.7% and an amino acid identity (of the available 3Dpol region) of 95.7%. Subsequently, Knowles and Wadsworth (2010) determined the complete genome sequence of the harbour seal picornavirus. The two viruses shared 84% and 92.3% nt identity in their 5’ and 3’ UTR’s, respectively. Their 3Dpol regions shared 87.2% nt and 98.6% amino acid (aa) identity while their VP1 regions shared 81.2% nt and 99.6% aa identity. This suggests that, not only do both seal picornaviruses belong to the same virus species, but also to the same antigenic type. The harbour seal’s range is limited to temperate and Arctic marine coastlines of the Northern hemisphere. They are found in coastal waters of the northern Atlantic and Pacific Oceans as well as those of the Baltic and North Seas, making them the most wide-ranging of the pinnipeds. The range of ringed seal’s is throughout the Arctic Ocean including the Baltic Sea, the Bering Sea and the Hudson Bay. The ranges of the two seal species overlap in the costal waters of Scandinavia. The close relationship between these two viruses suggests picornaviruses may circulate between different seal species. The role of picornaviruses in diseases of seals remains to be determined as does the broadness of their host specificity. Interestingly, both isolates of SePV-1 appear to possess two VPg's:
Figure: Predicted amino acid sequences of the two putative VPg’s. Black background, conserved residues between both viruses and both VPg’s. Grey background, conserved residues between both viruses for each VPg. This is the only picornavirus (apart from foot-and-mouth disease virus and the newly described mosavirus) so far found which potentially has multiple genome-linked proteins.A picornavirus, which is most closely related to seal aquamavirus 1 (SeAV-1), has been found in an Asian black bear (Ursus thibetanus) (Wang et al., 2018). The amino acid identity relationships between bear aquamavirus 1 (BeAV-1) and SeAV-1 are 87% (P1), 75% (P2) and 79% (P3), while the complete genome nucleotide identity is 71.7% (Wang et al., 2018). These figures suggest that BeAV-1 should belong to the species Aquamavirus A, however, it has been suggested by the authors that it might form a novel species within the genus Aquamavirus (Wang et al., 2018). Like seal aquamavirus 1, bear aquamavirus 1 appears to possess two VPg polypeptides. Genome organisation: VPg+5'UTRIRES-IVB[1AB-1C-1D-2Anpgp/2A-2B-2C/3A-3B1VPg1-3B2VPg2-3Cpro-3Dpol]3'UTR-poly(A) References Kapoor, A., Victoria, J., Simmonds, P., Wang, C., Shafer, R.W., Nims, R., Nielsen, O. and Delwart, E. (2008). A highly divergent picornavirus in a marine mammal. J. Virol. 82: 311-320. Knowles, N.J. (2005). A pan-picornavirus RT-PCR: identification of novel picornavirus species. EUROPIC 2005: XIIIth Meeting of the European Study Group on the Molecular Biology of Picornaviruses, Lunteren, The Netherlands, 23-29th May 2005. Abstract A06. Knowles, N.J. and Wadsworth, J. (2010). The complete genome sequence of a picornavirus isolated from a harbour (common) seal (Phoca vitulina). EUROPIC 2010: XVI Meeting of the European Study Group on the Molecular Biology of Picornaviruses, St. Andrews, Scotland, 11-16 September 2010. Abstract H16, p. 148. Osterhaus, A.D.M.E. (1988). Seal death. Nature, Lond. 334: 301-302. Wang H, Zhang W, Yang S, Kong N, Yu H, Zheng H, Gao F, Tong W, Li L, Wang X, Deng X, Delwart E, Shan T. Asian black bear (Ursus thibetanus) picornavirus related to seal aquamavirus A. Arch Virol. 2018 Dec 20. doi: 10.1007/s00705-018-4101-6. [Epub ahead of print] PubMed PMID: 30569277. |